introduction:
The winner of the 2016 Nobel Prize in Physiology and Medicine was Japanese scientist Yoshinori Ohsumi to reward him for his discovery of the mechanism of autophagy. What is "cell autophagy"? Autophagy is the basic process of degradation and reuse of cellular components. The word "autophagy" is derived from the Greek prefix "auto-" and another Greek word "phagein", the former meaning "self" and the latter meaning "swallowing." Therefore, the meaning of autophagy is very clear, that is, "self-phagocytosis" (Reference 1).
The concept of "autophagy" first appeared in the 1960s, when researchers discovered that cells can destroy their own internal matter by wrapping them into a membrane structure, forming small capsules and transporting them to what is called "solvent." The recovery mechanism of the enzyme body is decomposed. The study of “autophagy†was very difficult. Until the 1990s, after a series of excellent experiments, Japanese scientists used the baker's yeast to find key genes related to autophagy. He then began to work on the underlying mechanisms of autophagy in yeast, and found that similar complex processes exist in our human cells. The study of Otsuka's Good Book renewed our old view of the cellular material cycle, and his research opens up new paths to understanding the key role of autophagy in many physiological processes, such as the adaptation of organisms to hunger or the body's response to infection. Mutations in autophagy genes can lead to disease, and autophagy plays a role in some types of diseases such as cancer and neurological diseases (Reference 1).
In the latest research literature on "cell autophagy", we selected five articles, using "IncuCyte real-time dynamic cell imaging technology" to study autophagy from different aspects.
Essen BioScience's IncuCyte long-term dynamic live cell imaging and data analysis system enables real-time dynamic live cell imaging of up to six 384-well plates in an incubator at pre-set time points. IncuCyte ZOOM provides high-resolution phase contrast images, green fluorescence images, and red fluorescent images. Powerful analysis software allows quantitative analysis of images and curves and charts. It is suitable for cell proliferation monitoring, reporter gene, apoptosis, cytotoxicity, cell migration/invasion, angiogenesis, nerve cell tracking, chemotaxis, phagocytosis, immune cell killing and other applications. For specific introduction, please refer to:
IncuCyte introduces the URL link:
Http://?equipid=94008&division=405
The literature is briefly described as follows:
(A) Autophagy Supports Breast Cancer Stem Cell Maintenance by Regulating IL6 Secretion by regulating IL6 secretion (Reference 2, Impact Factor 4.510)
Literature Abstract:
"Cell autophagy" is the function of cells to survive in a stressful environment by degrading intracellular material to provide nutrients and energy. Autophagy is considered to be a key process for the maintenance of cancer stem cells (CSCs) or tumor initiating cells. But the mechanism by which autophagy supports CSC survival is still unknown. In the present study, autophagy was inhibited by knocking out the ATG7 or BECN1 gene, which inhibits the CD44 + /CD24 low/- group in breast cancer cells by modulating the secretion of CD24 and IL6. In breast cancer cell lines that do not depend on autophagy survival, autophagy inhibition increases the amount of IL6 secreted into the medium; whereas in autophagy-dependent cell lines, autophagy inhibition reduces IL6 secretion, cell survival, and Formation of microspheres (mammosphere). In these cells, IL6 treatment or provision of conditioned medium to cells with autophagy ability can rescue microspheres due to inhibition of autophagy. These results reveal that autophagy can regulate CSC maintenance in autophagy-dependent breast cancer cells by modulating IL6 secretion, thereby revealing that autophagy is a potential therapeutic target for breast cancer.
experimental method:
To detect cell proliferation and death, cells were seeded in 96-well plates and imaged using the IncuCyte system (Essen BioSciences). After the first scan, add the apoptotic Caspase 3/7 Green Detection Reagent or YOYO3 Reagent. After culturing for 72 hours, YOYO3 reagent was added. The cell growth curve was calculated from the confluence, and the cell death curve expressed the percentage of green/red fluorescence confluence. These data were obtained every 4 h using IncuCyte's 10x objective. In a mammosphere experiment, cells were seeded in 24-well plates. IncuCyte's full-hole imaging mode was used to shoot for 72h. The results show the percentage of red fluorescence convergence.
Experimental results:
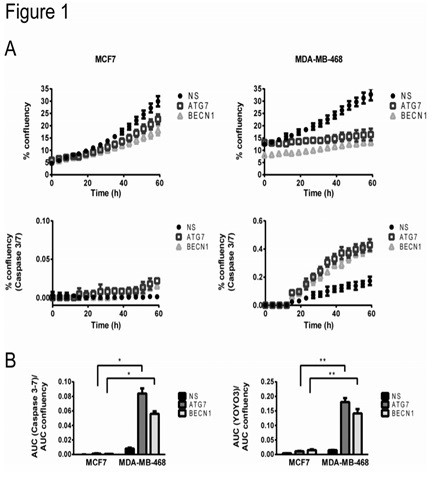
Figure 1. Cell proliferation and apoptosis of breast cancer cell lines that are dependent on autophagy and autophagy independent.
In Fig. 1, autophagy-independent MCF7 cells and autophagy-dependent MDA-MB-468 cells were transfected with lentiviruses containing shRNAs of the autophagy key gene ATG7 or BECN1 to knock out autophagy genes. NS indicates Non-silencing control, and ATG7 or BECN1 silencing indicates that autophagy is inhibited. Cellular proliferation (% of real-time image confluence) was detected using the IncuCyte imaging system, caspase activation (% confluency of Caspase 3/7 substrate emitting green fluorescence) and total cell death (YOYO3 red fluorescence in the same image) Percentage of confluence). In Figure A, a representative curve is shown for every three replicates. When autophagy was inhibited, in MDA-MB-468 cells, cell proliferation was significantly reduced and apoptosis was significantly increased compared with MCF7 cells. In panel B, the area under the curve (AUC) of the green confluency (caspase 3-7 activity, indicating apoptosis) curve, or the area under the curve (AUC) of the red confluency (total cell death expressed by YOYO3) curve. Normalization was performed using the AUC of the proliferation curve (representing the total number of cells). Comparison of autophagy-deficient MDA-MB-468 cells (expressing ATG7 or BECN1 shRNAs) and MCF7 cells showed significant apoptosis in MDA-MB-468 cells compared to MCF7 cells at autophagy inhibition (left panel) And cell death (right). In Panels A and B above, MCF7 cells that are not dependent on autophagy are not affected by autophagy inhibition.
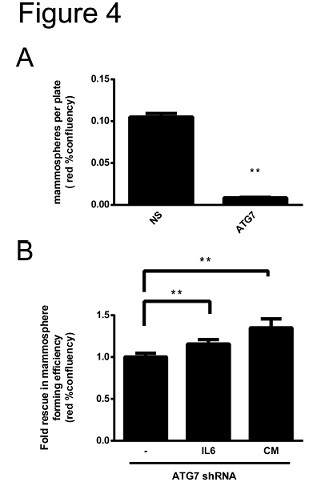
Figure 2. Autophagy-regulated secretion of IL6 contributes to microsphere formation in the MDA-MB-468 breast cancer cell line.
In Fig. 2, the nuclear-expressing cherry-fluorescent MDA-MB-468 cells were transfected with non-silencing (NS) shRNA or ATG7 shRNA, and the formation efficiency of the microspheres was evaluated by IncuCyte photographing. Figure A shows shooting for 7 days, and Figure B shows shooting for 48 hours. In panel A, autophagy inhibition results in a decrease in microsphere formation compared to control. In panel B, cells were either treated with IL6 at the time of planting or cultured using conditioned media (CM) obtained from non-transfected cells that had successfully produced microspheres for 3 days. Both treatments contribute to the formation of microspheres in the MDA-MB-468 breast cancer cell line.
(B) autophagy inhibition can promote the chemotherapy sensitivity of BRAF V600E brain tumor (Autophagy Inhibition Improves Chemosensitivity in BRAFV600E Brain Tumors) (Reference 3, impact factor 19.453)
Literature Abstract:
Autophagy suppression is a potential therapeutic strategy for cancer, but it is not known which tumors can be treated. Mutations in BRAF V600E have been shown to be important in pediatric central nervous system (CNS) tumors and can also affect autophagy behavior in other types of tumors. We evaluated CNS tumor cells with BRAF V600E mutation and found that mutant cells (non-wild type) showed high probability of induced autophagy and were sensitive to drug or gene-induced autophagy inhibition, and autophagy inhibition in clinical use. The combination of chloroquine and the Raf inhibitor vemurafenib, or chloroquine, showed synergy when used in combination with standard chemotherapy. Importantly, we have also demonstrated that chloroquine enhances the sensitivity of vemurafenib in resistant primary cultures in vitro and demonstrates for the first time that the addition of chloroquine to patients with V600E mutations with vemurafenib may increase clinical outcomes. These findings indicate that CNS tumors with BRAF V600E mutations are autophagy-dependent and can target autophagy inhibition in combination with other therapeutic strategies.
experimental method:
Tumor cell growth monitoring was performed using IncuCyte. The cells were seeded in 96-well plates at a density of 2000 per well and cultured with IncuCyte ZOOM for monitoring. Each well was imaged with 4 independent fields using a 10x objective, with an imaging interval of 4 hours. Three replicates were performed for each experiment, and the growth curve of the cells was shown by the degree of confluence.
Experimental results:

Figure 3. Cell growth curve of three cells
In Figure 3, panels (C) expressing control, ATG5 shRNAs and ATG12 shRNAs were grown in standard medium for 72 hours. Cell growth curves were obtained using an IncuCyte real-time dynamic imaging system. Data show mean ± SEM. The results showed that cells transfected with BRAF V600E mutants that knocked out ATG5 and ATG12 genes by shRNA reduced cell proliferation compared to the control group because gene knockdown inhibited autophagy.
(3) Autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells (Ref. 4, Impact Factor 8.459) )
Literature Abstract:
The aggressiveness of glioblastoma multiforme (GBM) is manifested in its invasion in situ and resistance to treatment. Among the formed GBMs, there is a type of tumor-forming cells (GBM stem cells, GSCs) having stem cell characteristics, which are thought to cause resistance to treatment. The metabolic pathway of autophagy is shown to be regulated by the survival of GBM. However, the autophagy state of GBM and its role in cancer stem cells have not been identified. We found that in GBM tumors carrying the mesenchymal signature, some autophagy regulators are highly expressed and can influence their aggressiveness and invasiveness, which is also related to the composition of the MAPK pathway. This autophagy marker includes the autophagy-related genes DRAM1 and SQSTM1, encoding a key selective autophagy regulator p62. Highly expressed DRAM1 is associated with a shorter survival of GBM patients. In GSCs, expression of DRAM1 and SQSTM1 is associated with activation of MAPK and expression of the mesenchymal cell marker c-MET. Knockout of DRAM1 reduced the localization of p62 on autophagosomes and its autophagy-mediated degradation, suggesting a role for DRAM1 in p62-mediated autophagy. In contrast, autophagy induced by starvation or mTOR/PI-3K inhibition was not affected by DRAM1 or p62 down-regulation. Functionally, DRAM1 and p62 regulate the movement and invasion of cells in GSCs. This is related to changes in energy metabolism, especially reduced ATP and lactate levels. Taken together, these findings elucidate the role of autophagy in GBM and reveal new functions of autophagy regulators DRAM1 and p62 in controlling migration/invasion of cancer stem cells.
experimental method:
Because autophagy was induced in active invasive/migrating glioblastoma stem cells (GSCs), we decided to determine whether knocking out autophagy DRAM1 and p62 would affect cell migration and/or invasion. Imaging and analysis were performed using the Migration Module of the IncuCyte Real-Time Dynamic Imager, and the curve of scratch healing over time was recorded. The horizontal axis represents time and the vertical axis represents the distance of cell movement.
Experimental results:
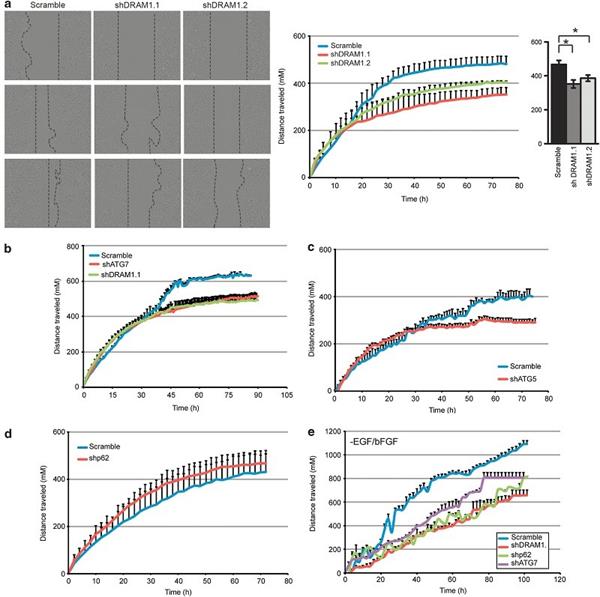
Figure 4. Effect of autophagy inhibition on cell migration
Figure 4 shows that down-regulation of autophagy regulators impairs cell migration induced by scratch assays and cell invasion by cell invasion assays. (a) Representative images of scratch assays using Scramble, shDRAM 1.1 and shDRAM 1.2 G166 cells. Images were acquired and analyzed every 2 hours via the IncuCyte Real Time Dynamic Imager. The scratch healing curve of the middle image showed cell migration for up to 78 hours. The image on the right shows the migration distance completed in 72 hours. The two shRNA sequences of DRAM1 significantly reduce the healing of scratches compared to Scramble cells. (b, c) knockdown of ATG7 and ATG5 inhibited migration, and in (b), the superimposed curve showed that knocking out ATG7 and knocking out DRAM1 had similar effects on cell migration. Knock-out p62(d) did not show any effect on the scratch migration experiment, probably because the growth ability of cells lacking p62 was improved, and cell growth was a distracting factor in the scratch test, so we also examined the removal of growth factors ( Scratch healing experiments of G166 cells of GF). (e) In GSCs lacking GF, down-regulation of p62 resulted in a similar reduction in scratch healing similar to knockout of DRAM1 and ATG7. Knockout of autophagy regulators impairs the motility of G166 cells lacking growth factors.
(Iv) by inhibiting the activation of mTORC1 MnK / eIF4E protein translation pathway promotes Chikungunya (Inhibition of mTORC1 Enhances the Translation of Proteins via Chikungunya the Activation of the MnK / eIF4E Pathway) ( Ref. 5, impact factor 7.562)
Literature Abstract:
Chikungunya virus (CHIKV), a major infectious disease path across five continents, is a positive-stranded mRNA that replicates using the cell's cap-dependent translation machinery. Although viral infection inhibits mTOR, a metabolic sensor that controls cap-dependent translation, viral proteins are efficiently translated. Treatment with Rapalog, or silencing of the mtor or raptor gene without silencing the rictor gene, further promotes CHIKV infection in cultured cells. Using biochemical experiments and real-time imaging techniques, we demonstrate that this effect is not related to autophagy or to type 1 interferon production. To provide in vivo experimental evidence relevant to our findings, we treated mice with mTORC1 inhibitors, showing increased lethality and showing greater sensitivity to CHIKV. A systematic review of the viral life cycle has shown that inhibition of mTORC1 has a particularly positive effect on viral proteins and can promote viral replication by increasing translation of both structural and non-structural proteins. Molecular analysis defines the role of phosphatidylinositol 3-kinase (PI3K) and MAP kinase-activated protein kinase (MnKs) activation, leading to hyperphosphorylation of eIF4E. Finally, we show that in the context of CHIKV inhibition of mTORC1, viral replication takes precedence over host translation by similar functions. Our study reveals an unexpected bypass pathway through which CHIKV protein translation overcomes viral-induced mTORC1 inhibition.
experimental method:
The efficiency of cells in the experiment by CHIKV infection was measured using IncuCyte's real-time imaging method. Mouse embryonic fibroblast cells (MEFs) were infected with CHIKV-GFP 5' in 24-well and 96-well plates, and the reporter gene was imaged using the IncuCyte real-time imaging system. Four independent 950 x 760 μm 2 regions in each well were imaged at 1 hour intervals using a 20x objective. Cultures were maintained at 37 ° C and cultured in Thermo's Hera cell 240 incubator, all three replicates. GFP+ cells were counted using IncuCyte software and the results were expressed as the number of green fluorescent subjects per mm 2 . The values ​​in the four fields of view in each well were summed and the average was calculated in conjunction with the three replicates. IncuCyte's experimental results can also be made into video.
Experimental results:
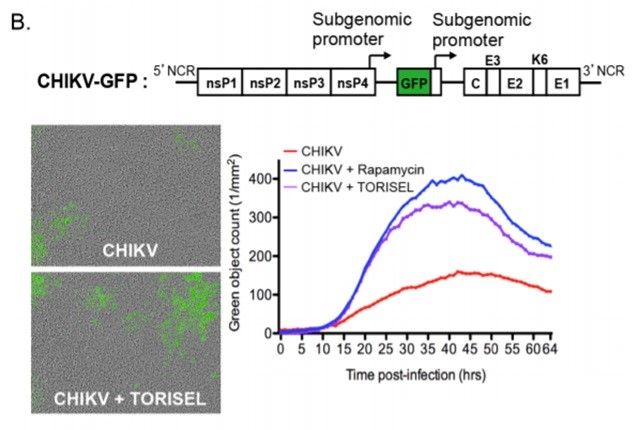
Figure 5. MEFs were infected with CHIKV-GFP and the number of GFP positive cells (reporter genes) in the cells was observed with IncuCyte.
Figure 5 (B) is a schematic diagram of the structure of CHIKV-GFP. MEFs lacking autophagy key genes were infected with CHIKV-GFP (MOI=1), and Rapamycin (100 nM) or TORISEL (0.1 mg/ml) was added. Both Rapamycin and TORISEL were inhibitors of mTORC1. GFP-positive cells were counted using a real-time dynamic imager, IncuCyte, which represents CHIKV-infected cells. The images show GFP staining (number of green subjects) and curves showing the dynamic process of infection, and similar results are shown in seven other independent experiments. The results showed that inhibition of mTOR promoted CHIKV infection in cultured cells, indicating that inhibition of mTORC1 promoted translation of viral proteins, but not autophagy.
(A) ABT-888 enhances cytotoxic effects of temozolomide independent of MGMT status in serum free cultured glioma cells in serum-free cultured glioma cells (ABT-888 enhances cytotoxic effects of temozolomide independent of MGMT status in serum free cultured glioma cells) Reference 6, impact factor 3.694)
Literature Abstract:
BACKGROUND: Current treatment criteria for Glioblastoma Multiforme (GBM) include fractionated focused radiotherapy and simultaneous temozolomide (TMZ) chemotherapy. A promising therapeutic strategy to increase the efficacy of TMZ is to interfere with DNA damage repair functions with poly ADP ribose polymerase protein inhibitor (PARPi). The goal of the current study is to explore the results of a combination of glioma stem-like cells (GSC) derived from patients.
Methods: The feasibility of combination therapy was tested on established GBM cell lines U373 and T98. We developed an in vitro drug screening method based on GSC cultures derived from tissue samples from primary patients (n=20) to assess the effects of PARPi (ABT-888) alone and the combination with TMZ. The therapeutic effects were tested by survival, double-strand break, apoptosis and autophagy, and longitudinal microscopic cell monitoring (IncuCyte) experiments were performed. The state of O-6-methylguanine-DNA methyltransferase (MGMT) was determined by methylation experiments and protein expression experiments of western blots.
Experimental results: Individual treatment with 10 μM PARPi reduced cell survival by more than 25% with a probability of 4 (20%) of 20 GSCs. Individual treatments of 50 μM and 100 μM TMZ were effective for 12 and 14 of 20 GSCs, respectively. In the 100 μM dose, TMZ resistance was found in 7 out of 8 MGMT protein positive cultures. The synergy between TMZ treatment and PARPi was found in 90% of GSCs (n=20), 6 of which were initial resistance and 7 were sensitive to TMZ alone. Increased double-strand break induction and apoptosis were also observed in responding GSCs. Although not statistically significant, we have noted a trend in which autophagy is observed in both western blots and autophagy.
CONCLUSION: When performed simultaneously, PARPi-mediated synergy of TMZ is independent of TMZ sensitivity and can reverse MGMT(-)-mediated resistance. The response to combination therapy is accompanied by increased induction of double-strand breaks, consistent with increased apoptosis and autophagy. The addition of PARPi synergizes with TMZ treatment in primary GSCs. PARPi can potentially increase the effectiveness of GBM standard treatment.
experimental method:
Cell proliferation assay: Cell confluence was observed using an IncuCyte real-time dynamic live cell imager. The cells were seeded in 96-well plates and placed in IncuCyte. A phase difference image was taken every 1-3 hours, and each well was calculated using IncuCyte software. Convergence.
Double-strand break assay: The number of yH2Ax positive cells was observed with IncuCyte.
Apoptosis: To detect apoptosis induced by treatment alone or in combination therapy, add Essen BioScience's Cell Player Caspase 3/7 Apoptosis Reagent at the same time as the drug is added, the dilution ratio is 1:1000, and the reagent is recorded by IncuCyte FLR. The ratio of apoptotic cells to total cells after 48 hours.
Autophagy: To detect autophagy induced by separate therapy and combination therapy, we used Enzo Life Sciences' Cyto-ID autophagy test kit, which provides a single dansyl sulfonylamine dye, specifically The phagosome is stained. The cells were treated with the drug and observed with IncuCyte FLR to detect autophagosomes. In the software processing process, the fluorescence intensity threshold of the autophagosome is first set, and the number of autophagosomes per well is calculated.
Experimental results:
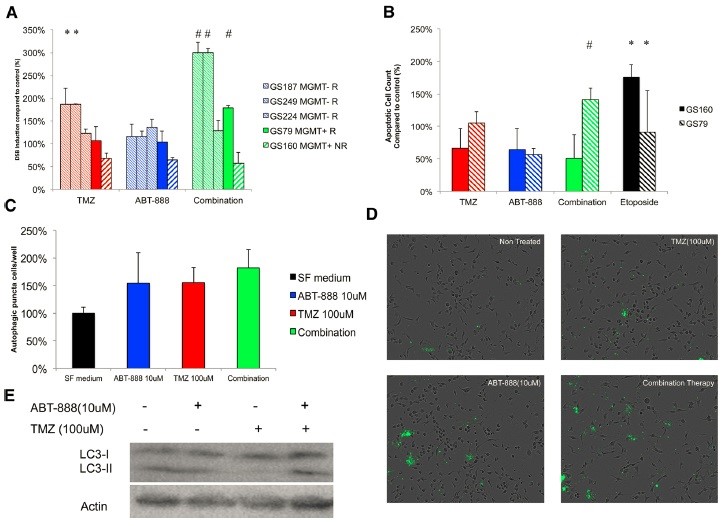
Figure 6. PARP inhibition increases TMZ-induced double-strand breaks in GSCs, autophagy and apoptosis.
Figure 6 shows that (A) combination therapy promotes DSB induction. The number of yH2Ax positive cells in GSCs, MGMT(+) and MGMT(-) was calculated as an indicator of DSB induction after 16 hours of drug treatment. The results of GS160 cells were quite significant compared to the non-treated control group. (B) Combination therapy significantly promoted apoptosis induction in response to GSCs without promoting non-responsive GSCs. Real-time imaging was used to calculate the number of cells positive for apoptosis after 48 hours of drug treatment. After combination therapy, apoptotic cells of GSC79 cells were significantly increased compared to treatment alone. GS160 cells were not affected by the combination of TMZ/ABT-888 drugs, while positive controls were able to successfully induce apoptosis. (C) TMZ and ABT-888 can induce autophagy in GS79 cells, and combination therapy increases this effect. The number of autophagosomes in each well was calculated and compared to the untreated control. The drug was read after 16 hours. The number of all autophagosomes treated increased compared to the control. However, the combination therapy was not significantly increased compared to treatment with ABT-888 or TMZ alone. (D) Representative images obtained from IncuCyte FLR reflect GS79 stained autophagosomes. Note the accumulation of fluorescent spots inside the cells, especially those with enlarged lesions. (E) In GS79, combination therapy promoted autophagic flux compared to treatment alone. Western blotting was performed 24 hours after the addition of the drug, indicating accumulation of LC3-11, indicating an increase in autophagic flux. Actin is a loading control group.
to sum up:
Representative literatures from the above five autophagy studies were performed using the IncuCyte long-term dynamic live cell imaging and data analysis system of Essen BioScience. To learn more about IncuCyte instruments and real-time dynamic live cell imaging technology, please contact Essen's China distributor, Shanghai Dian Biotech Co., Ltd., for contact information, see the “Request for Information†section below the article.
references:
(1) What is the “cell autophagy†that won the Nobel Prize: an important mechanism for human cells, Sina Technology Weibo, October 3, 2016
(2) Maycotte, P. et.al. , Autophagy Supports Breast Cancer Stem Cell Maintenance by Regulating IL6 Secretion, Mol Cancer Res. , 2015
(3) Mulcahy Levy JM et.al. , Autophagy Inhibition Improves Chemosensitivity in BRAF V600E Brain Tumors, Cancer Discov. 2014 July, 4(7), pp.773-780
(4) Galavotti, S. et.al. , The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells, Oncogene , 2013, 32, pp. 699-712
(5) Joubert, PE et.al. Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins via the Activation of the MnK/eIF4E Pathway, PLOS Pathogens , 2015, August 28
(6) Balvers, R. et.al. , ABT-888 enhances cytotoxic effects of temozolomide independent of MGMT status in serum free cultured glioma cells, Journal of Translational Medicine , 2015, 13:74
Security Camera
Security Camera Surveillance camera is a semiconductor imaging device, which has the advantages of high sensitivity, strong light resistance, small distortion, small size, long life, and anti-vibration. Surveillance cameras in security systems.
Image generation is currently mainly from CCD cameras, and the stored charge can also be taken out to change the voltage. It has the characteristics of anti-vibration and impact and is widely used.
Security Camera Surveillance camera is a semiconductor imaging device, which has the advantages of high sensitivity, strong light resistance, small distortion, small size, long life, and anti-vibration. Surveillance cameras in security systems.
Image generation is currently mainly from CCD cameras, and the stored charge can also be taken out to change the voltage. It has the characteristics of anti-vibration and impact and is widely used.
Security Camera Surveillance camera is a semiconductor imaging device, which has the advantages of high sensitivity, strong light resistance, small distortion, small size, long life, and anti-vibration. Surveillance cameras in security systems.
Image generation is currently mainly from CCD cameras, and the stored charge can also be taken out to change the voltage. It has the characteristics of anti-vibration and impact and is widely used.
Security Camera,CCTV Camera,Solar Security Camera ,4G Security Camera
Shenzhen Fuvision Electronics Co., Ltd. , https://www.outdoorsolarcamera.com