Gene editing technology refers to the editing of target genes to achieve knockout and insertion of specific DNA fragments. Since the publication of the CRISPR/Cas9 gene editing technology, a series of major breakthroughs have been made, and one of the top ten scientific advances has been ranked by Science magazine in 2012, 2013, 2015 and 2017. Therefore, CRISPR/Cas9 has been favored by many researchers and investors for its ease of operation and low cost.
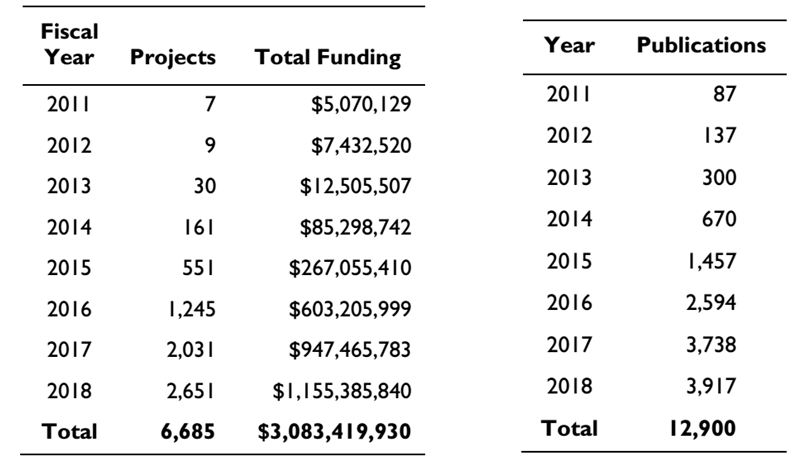
As shown in the following figure: From 2011 to 2018, NIH's funding for CRISPR-related research has grown rapidly from more than $5 million to $1.1 billion, and the number of CRISPR-related scientific publications has grown dramatically from the initial 87 to 3,917. These data reflect the huge potential value of CRISPR / Cas9.
In 2018, the CRISPR system continued to exert its strength and made breakthrough progress in many fields. In this issue, we will first sort out relevant major events from the aspects of CRISPR system development and mechanism research.
I. The new CRISPR system continues to expand the scope of genome editing. Since the earliest published spCas9 in 2013 [1], scientists have been working to find more homologs of Cas9 in complex bacterial populations to expand the library of gene editing tools. Overcoming many problems in the existing Cas9 system, such as the component is too large to be packaged into AAV (adeno-associated virus vector), PAM can not cover the entire genome and so on. With the advent of saCas9, Cpf1 and Cas13 (pictured), the new gene editing system was discovered. In addition to editing DNA, it gradually began to have more functions such as editing RNA and single-stranded nucleotides. The discovery of these new systems not only expanded. The editing scope of CRISPR also extends its application in many fields. In 2018, more potential CRISPR systems were identified and transformed by scientists.

At the end of 2017, the Zhang Feng team first reported the discovery of the Cas13 system, which confirmed its ability to target RNA in mammalian cells [2].
On March 15, 2018, Konermann and others at the Salk Institute added a new member to the Cas13 family: Cas13d. This is a CRISPR/Cas system from the gut bacteria (L. vaginalis XPD3002), named CasRx. Similar to Cas13, CasRx specifically targets and cleaves RNA, but is 20% smaller than other Cas13s. At the same time, CasRx-mediated knockdown is more efficient and specific than other RNA regulation methods [3].
2. nuclease cyclic (ring nuclease): virus defense termination state "switch"
Type III effector complexes have recently been shown to bind to the genetic material of invading viruses to form a circular oligoadenylate, commonly known as the second messenger, which binds and activates ribonuclease and other proteins via the CRISPR-associated Rossman folding domain. Factor to resist the invasion of the virus, making the cells into an anti-viral state. However, the continued activation of this state is detrimental to the cell, and the researchers speculate that there may be some mechanism to turn this state off after the virus is cleared. On September 19, 2018, the Malcolm White team confirmed this mechanism, a protein called a circular nuclease specifically cleaves cyclic oligoadenylation, thereby terminating the antiviral state. The identification of circular nucleases has increased the understanding of the CRISPR system [4].
3. Cas14: The smallest Cas protein at present, adding a weapon for disease diagnosis
On October 18, 2018, the Jenifer Doudna team discovered the smallest functional CRISPR system to date, the Cas14. Cas14 is only 400-700 amino acids, but like Cas12 and Cas13, it can target single-stranded DNA (ssDNA) without the restriction sequence requirements, thus blindly cutting all ssDNA in cells. This feature makes high-fidelity single nucleotide polymorphism genotyping possible. With further improvements, more options are available for the existing diagnostic system (DETECTR) [5].
4. Cas12 protein new team member, further expand the toolbox of the CRISPR system
Cas12a (Cpf1) and SpCas9 have been used as the most commonly used CRISPR-Cas gene editing tools and have been successfully applied in various fields of genome engineering. Compared to SpCas9, Cas12a is widely studied and used with its small volume (1228 bp) and the guidance of only a single RNA [6]. On December 6, 2018, researchers from Arbor Biotechnologies in the United States discovered a new group of Cas12 protein members: Cas12c, Cas12g, Cas12h, and Cas12i. Among them, Cas12c, Cas12h and Cas12i have RNA-directed double-strand DNA cleavage activity, and it was also found that Cas12i showed significant difference in cleavage efficiency on the complementary and non-complementary strands of the CRISPR crRNA spacer, which resulted in the formation of dsDNA mainly Chain nicking. Cas12g cleaves single-stranded RNA and single-stranded DNA by RNA targeting mainly in the form of ribonuclease. This study reveals the functional diversity of the V-type CRISPR-Cas system in the evolution of different routes, and further expands the scope of application of the CRISPR toolbox [7].
Second, the engineering of CRISPR systems to expand the scope of genomic applications <br> Although the CRISPR-Cas9 system is widely used for genome editing, the range of sequences that Cas9 can recognize is limited by the specific original spacer neighboring motif (PAM) requirements, so usually It is difficult to achieve high-precision genome-editing applications for targeting double-stranded DNA breaks, including popular single-base editing and CRISPR-based gene screening. With the in-depth study of Cas9 proteomics, it is possible to break this restriction by artificially introducing random mutations to alter PAM-specific Cas9 derivatives. At present, scientists mainly identify Cas9 functional mutants of different PAM sequences through structural information and directed evolution-based bacterial selection systems. These Cas9 mutants have comparable editing ability and specificity to wild-type SpCas9. This technology provides a research direction for finding high-precision Cas9 mutants, greatly expanding the CRISPR system toolkit, and opening the door for the whole genome editing of the CRISPR system [8].
1. Establish a new bacterial defense system screening system to develop more molecular tools of potential value
On January 25, 2018, the Rotem Sorek team from the Weizmann Institute of Science in Israel published a paper on Science to develop more defensive system genes by constructing a computer program that scans all bacterial genomes. The system is inserted into bacteria that have been inactivated by the natural immune system. Through the screening of phage and other infectious agents, it is found that there are 10 previously unknown bacterial immune defense mechanisms. Any of these new defense systems is likely to be the next gene editing tool [9].
2. xCas9: a SpCas9 mutant that recognizes multiple PAM sequences
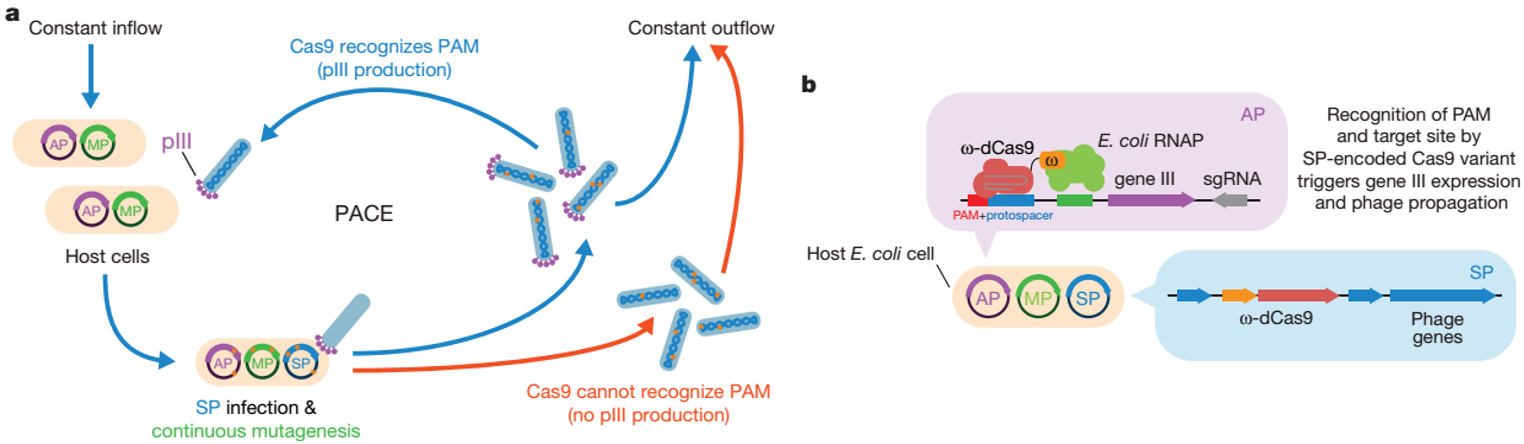
On February 28, 2018, the David Liu team published a journal in Nature, using a phage-assisted continuous evolution system (PACE) to evolve a variant of SpCas9 (xCas9) that recognizes multiple PAM sequences. The PAM compatibility of xCas9 is by far the most widely used in all Cas9 recognition mammals and can be applied to human cells, including targeted transcriptional activation, nuclease-mediated gene knockout, single base editing, etc. . It is worth noting that although the PAM compatibility of xCas9 is expanded, its DNA specificity is much higher than that of SpCas9, and the off-target effect of whole genome in NGG PAMs and non-NGG PAMs target sites is also lower than spCas9. many. The best xCas9 PAMs are NGN, accounting for about a quarter of the human genome. The emergence of xCas9 greatly expands the DNA targeting range of the CRISPR system, making the CRISPR system more accurate and flexible [10].
On September 21, 2018, the Osamu Nureki team developed a SpCas9 mutant (SpCas9-NG) that recognizes NG instead of NGG through rational design. SpCas9-NG increases the targeting range and has similar specificity to wild-type SpCas9 and can also be used with other editors (cytidine deaminase). Therefore, SpCas9-NG effectively complements the CRISPR toolbox and will play an important role in a wide range of applications from basic research to clinical treatment [11].
4. ScCas9: PAM using bioinformatics found containing only one Cas9 "G" of
On October 24, 2018, researchers from the Massachusetts Institute of Technology built an automated bioinformatics pipeline to further explore the neglected strains of the straight strain of Cas9 streptococcus by searching for PAMs (SPAMALOT). PAM diversity, a Cas9 (ScCas9) from Streptococcus mutans was discovered, demonstrating its ability to accurately edit in bacteria and human cells. The PAM sequence of ScCas9 is 5'-NNGTT-3' and contains only one base G, so it can target the target DNA sequence of SpCas9 and has more sites. ScCas9 can be used as an alternative genome editing tool or as a functional platform for the discovery of new Streptococcus PAM specificity [12].
Third, the progress of the mechanism of nuclease action in CRISPR system has made progress
Cas protein is a class of nucleases in the CRISPR/Cas system and at least 45 Cas protein families have been identified. Researchers have used cryo-electron microscopy to analyze their protein structure and mechanism of action, providing a basic structural and theoretical basis for understanding the application of gene editing systems and transforming CRISPR systems [13]. Now, a variety of new Cas proteins have been discovered and resolved, which will lay the foundation for the development of new CRISPR systems and their efficient use in gene therapy in the future.
1. Two studies reveal the importance of Cas4 nuclease for CRIPSR immune response
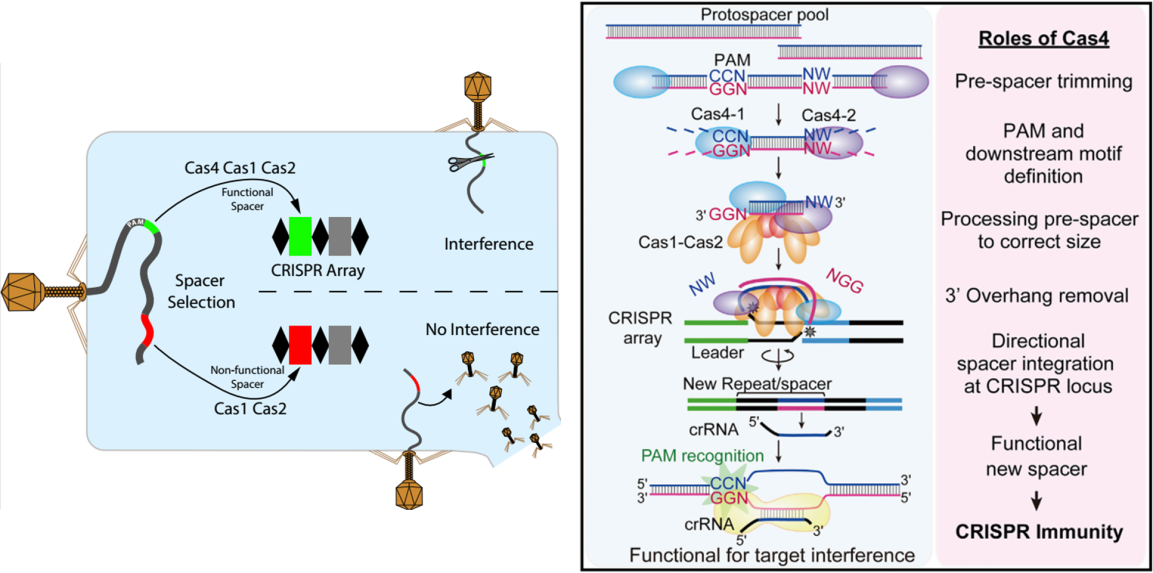
On March 27, 2018, the Shiimori team published a report in the Cell Reports journal, demonstrating that Cas4 nucleases are required for targeted selection of spacer sequence proximity motifs (PAMs), facilitating the selection of new CRISPRs by Cas1 and Cas2. The spacers are given to the natural host Synechocystis I-D CRISPR interference ability [14]. In the same year, the Shiimori team published a work in the Molecular Cell journal on June 7th, demonstrating that Cas4 immunoreacted with CRISPR-Cas by DNA sequence integration in the thermophilic archaea P. furiosus and identified two Cas4s ( Cas4-1 and Cas4-2) capture the DNA fragment of the P. furiosus CRISPR spacer and its key role in the processing of the original spacer. On the other hand, Cas4 ensures that the CRISPR spacers are integrated in a specific direction, ultimately triggering the CRISPR immune response [15]. Taken together, these findings provide a key role for Cas4 nucleases in CRISPR arrays, providing for in situ spacer generation and functional compartment integration.
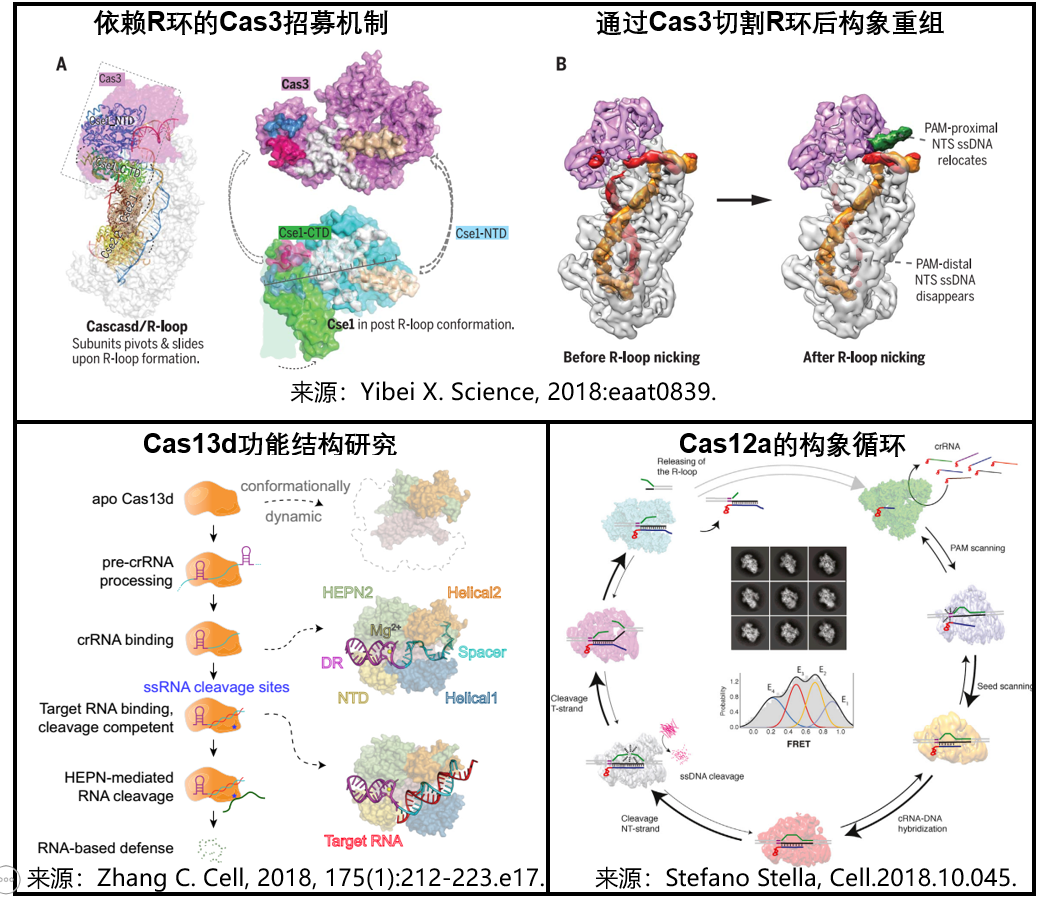
1) Interpretation of the molecular mechanism of the type I CRISPR/Cas system : On July 6, 2018, researchers from Cornell University and Harvard Medical School published a paper in the journal Science to study the use of single-particle cryo-electron microscopy (cryo-EM). Analysis of the structure of the TfuCascade/R-loop/Cas3 ternary complex before and after R-loop cleavage reveals the confusing Cas3 recruitment, DNA cleavage and degradation mechanisms. These studies provide a structural basis for understanding the molecular mechanism of action of the Type I CRISPR/Cas system [16].
2) Cas13d functional structure study : On September 20, 2018, scientists at the Salk Institute in the United States used cryo-electron microscopy to analyze the structure and a series of Cas13d-sgRNA binary complexes and Cas13d-sgRN-target RNA ternary complexes. The dynamic process, which allows us to see in more detail how the Cas13d-system is guided to RNA and the process of cutting. This has improved the CRISPR system for us to lay the foundation for the treatment of RNA diseases more effectively [17].
3) Cas12d loop conformation analytical: December 13, 2018, researchers from Novo Nordisk Foundation Center using a cryogenic electron microscopy, clarified Cas12a targeting DNA cleavage and degradation of ssDNA random mechanism, revealing Cas12a cleaves its target DNA, releasing non-specific cleavage activity, and degrading ssDNA molecules after activation. This allows us to adjust the CRISPR engine to achieve a specific desired effect [18].
The CRISPR/Cas gene editing system has great potential in human therapeutic applications. A full understanding of the repair mechanism of the in vivo editing system will help us to perform gene editing more efficiently and accurately, and provide sufficient theory and technology for safe and effective clinical applications. basis.
1. Fanconi anemia pathway plays a key role in the process of editing and forming DSBs in CRISPR-Cas9 editing
In August 2018, researchers at the University of California, Berkeley published a journal in the journal Nature Genetics, overturning the previous hypothesis that "cells repair genes after Cas9 enzyme cleavage of DNA". Researchers use CRISPR interference technology to 2000. Multiple genes were silenced and the FANCD2 protein in the Fanconi anemia pathway was localized to Cas9-induced DSBs, suggesting that it plays a key role in regulatory genome editing - regulation of FANCD2 protein can increase HDR frequency . At the same time, it was found that the Fanconi anemia pathway was not related to NHEJ, but the mechanism of repair to the single-chain template repair mechanism was improved by increasing the efficiency of homologous recombination. Therefore, the editing efficiency of HDR can be improved by regulating the activity of the Fanconi anemia pathway. This finding will help increase the efficiency of insertion of foreign DNA into the genome and ensure that CRISPR editing achieves the desired results [19].
On July 19, 2018, Wu Qiang's research team from Shanghai Jiaotong University published the latest research results of the team in the Molecular Cell journal, subverting the existing gene editing theory. They found that the cis-cutting of Cas9-cleaved DNA produced a prominent end, not just the previously reported flat-end end form, but also used high-throughput sequencing to analyze the DNA repair methods of paired-directed RNA editing and found that the repair of the broken ends It is predictable and presents a very regular form. This disruptive discovery laid a solid foundation for optimizing and transforming gene editing techniques [20].
Subsequently, on November 27, 2018, the Felicity Allen team also published a report in Nature. They synthesized more than 40,000 pairs of guided RNA and targeted DNA sequence editing library systems to explore the cutting and repair mechanism of CRISPR/Cas9, and clarified the results of the repair. Depending on the DNA sequence of the targeting region, most reproducible mutations are the result of single base insertions, short deletions or longer micro-homology-mediated deletions. At the same time, a machine learning algorithm (FORECasT) was developed using Big Data, which can be used to predict the corrected results using only the target site DNA sequence [22]. The in-depth understanding of these mechanisms and the development of new tools will help people better design genetic editing experiments.
In summary, we briefly introduced the important events in the development and mechanism research of the new CRISPR system in 2018. At present, CRISPR-Cas9 has become a simple, accurate and rapid gene editing technology, which is widely used in many life science related fields such as medicine, agriculture, forestry and animal husbandry, such as eye diseases, blood diseases and other genetic diseases. At the same time, we also recognize that this technology still has many problems and many potentials to be further researched and expanded. We believe that in the near future, CRISPR-Cas9 will show its invaluable value in various fields.
references:
1. 201302_Science_Multiplex Genome Engineering Using CRISPR-Cas Systems
2. Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13[J]. Science, 2017: eaaq0180.
3. Konermann, Silvana, et al. “Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors.†Cell (2018) pii: S0092-8674(18)30207-1.
4. Athukoralage JS, Rouillon C, Graham S, et al. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate [J]. Nature, 2018.
5. Lucas B. Harrington1,*,†, David Burstein2,*,‡, Janice S. Chen et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science, 2018.
6. Zetsche B, Gootenberg J, Abudayyeh O, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System [J]. Cell, 2015: S0092867415012003.
7. Winston X. Yan et al. Functionally diverse type V CRISPR-Cas systems, 2018, doi:10.1126/science.aav7271.
8. Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities[J]. Nature, 2015, 523(7561):481-485.
9. Doron S, Melamed S, Ofir G, et al. Systematic discovery of antiphage defense systems in the microbial pangenome [J]. Science, 2018: eaar4120.
10. Hu JH , Miller SM , Geurts MH , et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity [J]. Nature.
11. Hiroshi Nishimasu, Xi Shi, Soh Ishiguro et al. Engineered CRISPR/Cas9 nuclease with expanded targeting space. Science, 21 Sep 2018, 361(6408):1259-1262, doi:10.1126/science.aas9129.
12. Pranam Chatterjee1,2,*,*,terjee1,2,*,1,2,*,ered CRISPR/Cas9 nuclease with expanded targeting space. Science, 21 Sep 20ortholog. Science Advances, 24 Oct 2018, 4(10) :eaau0766.
13. Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation[J]. Science, 2014, 343(6176):1247997.
14. Sebastian N. Kieper, Cristóbal Almendros, Juliane Behler et al. Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell Reports, 27 March 2018, 22(13):3377–3384, doi:10.1016/j.celrep. 2018.02.103.
15. Masami S, Garrett SC, Graveley BR, et al. Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci [J]. Molecular Cell, 2018, 70(5): 814-824.e6 .
16. Yibei X, Min L, Dolan AE, et al. Structure basis for RNA-guided DNA degradation by Cascade and Cas3 [J]. Science, 2018: eaat0839.
17. Zhang C, Konermann S, Brideau NJ, et al. Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d [J]. Cell, 2018, 175(1): 212-223.e17.
18. Stefano Stella, Pablo Mesa, Johannes Thomsen, et al. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity, Cell (2018). DOI: 10.1016/j.cell.2018.10.045.
19. Chris D. Richardson, Katelynn R. Kazane, Sharon J. Feng et al. CRISPR–Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nature Genetics, August 2018, 50(8):1132–1139, doi :10.1038/s41588-018-0174-0.
20. Precise and Predictable CRISPR Chromosomal Rearrangements Reveal Principles of Cas9-mediated Nucleotide Insertion
21. Max W. Shen et al., Predictable and precise template-free CRISPR editing of pathogenic variants, Nature, 2018, DOI: 10.1038/s41586-018-0686-x.
22. Felicity Allen et al, Predicting the mutations generated by repair of Cas9-induced double-strand breaks, Nature Biotechnology (2018). DOI: 10.1038/nbt.4317.
As shown in the following figure: From 2011 to 2018, NIH's funding for CRISPR-related research has grown rapidly from more than $5 million to $1.1 billion, and the number of CRISPR-related scientific publications has grown dramatically from the initial 87 to 3,917. These data reflect the huge potential value of CRISPR / Cas9.

Figure 1. CRISPR research funding (left) and publications increase year by year (right) (source NIH)
In 2018, the CRISPR system continued to exert its strength and made breakthrough progress in many fields. In this issue, we will first sort out relevant major events from the aspects of CRISPR system development and mechanism research.
I. The new CRISPR system continues to expand the scope of genome editing. Since the earliest published spCas9 in 2013 [1], scientists have been working to find more homologs of Cas9 in complex bacterial populations to expand the library of gene editing tools. Overcoming many problems in the existing Cas9 system, such as the component is too large to be packaged into AAV (adeno-associated virus vector), PAM can not cover the entire genome and so on. With the advent of saCas9, Cpf1 and Cas13 (pictured), the new gene editing system was discovered. In addition to editing DNA, it gradually began to have more functions such as editing RNA and single-stranded nucleotides. The discovery of these new systems not only expanded. The editing scope of CRISPR also extends its application in many fields. In 2018, more potential CRISPR systems were identified and transformed by scientists.

Figure 2. Cas9 homologs reported in recent years
1. CasRx : A stronger version of the Cas13 system At the end of 2017, the Zhang Feng team first reported the discovery of the Cas13 system, which confirmed its ability to target RNA in mammalian cells [2].
On March 15, 2018, Konermann and others at the Salk Institute added a new member to the Cas13 family: Cas13d. This is a CRISPR/Cas system from the gut bacteria (L. vaginalis XPD3002), named CasRx. Similar to Cas13, CasRx specifically targets and cleaves RNA, but is 20% smaller than other Cas13s. At the same time, CasRx-mediated knockdown is more efficient and specific than other RNA regulation methods [3].
2. nuclease cyclic (ring nuclease): virus defense termination state "switch"
Type III effector complexes have recently been shown to bind to the genetic material of invading viruses to form a circular oligoadenylate, commonly known as the second messenger, which binds and activates ribonuclease and other proteins via the CRISPR-associated Rossman folding domain. Factor to resist the invasion of the virus, making the cells into an anti-viral state. However, the continued activation of this state is detrimental to the cell, and the researchers speculate that there may be some mechanism to turn this state off after the virus is cleared. On September 19, 2018, the Malcolm White team confirmed this mechanism, a protein called a circular nuclease specifically cleaves cyclic oligoadenylation, thereby terminating the antiviral state. The identification of circular nucleases has increased the understanding of the CRISPR system [4].
3. Cas14: The smallest Cas protein at present, adding a weapon for disease diagnosis
On October 18, 2018, the Jenifer Doudna team discovered the smallest functional CRISPR system to date, the Cas14. Cas14 is only 400-700 amino acids, but like Cas12 and Cas13, it can target single-stranded DNA (ssDNA) without the restriction sequence requirements, thus blindly cutting all ssDNA in cells. This feature makes high-fidelity single nucleotide polymorphism genotyping possible. With further improvements, more options are available for the existing diagnostic system (DETECTR) [5].
4. Cas12 protein new team member, further expand the toolbox of the CRISPR system
Cas12a (Cpf1) and SpCas9 have been used as the most commonly used CRISPR-Cas gene editing tools and have been successfully applied in various fields of genome engineering. Compared to SpCas9, Cas12a is widely studied and used with its small volume (1228 bp) and the guidance of only a single RNA [6]. On December 6, 2018, researchers from Arbor Biotechnologies in the United States discovered a new group of Cas12 protein members: Cas12c, Cas12g, Cas12h, and Cas12i. Among them, Cas12c, Cas12h and Cas12i have RNA-directed double-strand DNA cleavage activity, and it was also found that Cas12i showed significant difference in cleavage efficiency on the complementary and non-complementary strands of the CRISPR crRNA spacer, which resulted in the formation of dsDNA mainly Chain nicking. Cas12g cleaves single-stranded RNA and single-stranded DNA by RNA targeting mainly in the form of ribonuclease. This study reveals the functional diversity of the V-type CRISPR-Cas system in the evolution of different routes, and further expands the scope of application of the CRISPR toolbox [7].
Second, the engineering of CRISPR systems to expand the scope of genomic applications <br> Although the CRISPR-Cas9 system is widely used for genome editing, the range of sequences that Cas9 can recognize is limited by the specific original spacer neighboring motif (PAM) requirements, so usually It is difficult to achieve high-precision genome-editing applications for targeting double-stranded DNA breaks, including popular single-base editing and CRISPR-based gene screening. With the in-depth study of Cas9 proteomics, it is possible to break this restriction by artificially introducing random mutations to alter PAM-specific Cas9 derivatives. At present, scientists mainly identify Cas9 functional mutants of different PAM sequences through structural information and directed evolution-based bacterial selection systems. These Cas9 mutants have comparable editing ability and specificity to wild-type SpCas9. This technology provides a research direction for finding high-precision Cas9 mutants, greatly expanding the CRISPR system toolkit, and opening the door for the whole genome editing of the CRISPR system [8].
1. Establish a new bacterial defense system screening system to develop more molecular tools of potential value
On January 25, 2018, the Rotem Sorek team from the Weizmann Institute of Science in Israel published a paper on Science to develop more defensive system genes by constructing a computer program that scans all bacterial genomes. The system is inserted into bacteria that have been inactivated by the natural immune system. Through the screening of phage and other infectious agents, it is found that there are 10 previously unknown bacterial immune defense mechanisms. Any of these new defense systems is likely to be the next gene editing tool [9].
2. xCas9: a SpCas9 mutant that recognizes multiple PAM sequences
On February 28, 2018, the David Liu team published a journal in Nature, using a phage-assisted continuous evolution system (PACE) to evolve a variant of SpCas9 (xCas9) that recognizes multiple PAM sequences. The PAM compatibility of xCas9 is by far the most widely used in all Cas9 recognition mammals and can be applied to human cells, including targeted transcriptional activation, nuclease-mediated gene knockout, single base editing, etc. . It is worth noting that although the PAM compatibility of xCas9 is expanded, its DNA specificity is much higher than that of SpCas9, and the off-target effect of whole genome in NGG PAMs and non-NGG PAMs target sites is also lower than spCas9. many. The best xCas9 PAMs are NGN, accounting for about a quarter of the human genome. The emergence of xCas9 greatly expands the DNA targeting range of the CRISPR system, making the CRISPR system more accurate and flexible [10].

Figure 3. PACE-phage-assisted continuous evolution mode pattern [10]
3. SpCas9-NG : A Cas9 that recognizes the "NG" PAM sequence On September 21, 2018, the Osamu Nureki team developed a SpCas9 mutant (SpCas9-NG) that recognizes NG instead of NGG through rational design. SpCas9-NG increases the targeting range and has similar specificity to wild-type SpCas9 and can also be used with other editors (cytidine deaminase). Therefore, SpCas9-NG effectively complements the CRISPR toolbox and will play an important role in a wide range of applications from basic research to clinical treatment [11].
4. ScCas9: PAM using bioinformatics found containing only one Cas9 "G" of
On October 24, 2018, researchers from the Massachusetts Institute of Technology built an automated bioinformatics pipeline to further explore the neglected strains of the straight strain of Cas9 streptococcus by searching for PAMs (SPAMALOT). PAM diversity, a Cas9 (ScCas9) from Streptococcus mutans was discovered, demonstrating its ability to accurately edit in bacteria and human cells. The PAM sequence of ScCas9 is 5'-NNGTT-3' and contains only one base G, so it can target the target DNA sequence of SpCas9 and has more sites. ScCas9 can be used as an alternative genome editing tool or as a functional platform for the discovery of new Streptococcus PAM specificity [12].
Third, the progress of the mechanism of nuclease action in CRISPR system has made progress
Cas protein is a class of nucleases in the CRISPR/Cas system and at least 45 Cas protein families have been identified. Researchers have used cryo-electron microscopy to analyze their protein structure and mechanism of action, providing a basic structural and theoretical basis for understanding the application of gene editing systems and transforming CRISPR systems [13]. Now, a variety of new Cas proteins have been discovered and resolved, which will lay the foundation for the development of new CRISPR systems and their efficient use in gene therapy in the future.
1. Two studies reveal the importance of Cas4 nuclease for CRIPSR immune response
On March 27, 2018, the Shiimori team published a report in the Cell Reports journal, demonstrating that Cas4 nucleases are required for targeted selection of spacer sequence proximity motifs (PAMs), facilitating the selection of new CRISPRs by Cas1 and Cas2. The spacers are given to the natural host Synechocystis I-D CRISPR interference ability [14]. In the same year, the Shiimori team published a work in the Molecular Cell journal on June 7th, demonstrating that Cas4 immunoreacted with CRISPR-Cas by DNA sequence integration in the thermophilic archaea P. furiosus and identified two Cas4s ( Cas4-1 and Cas4-2) capture the DNA fragment of the P. furiosus CRISPR spacer and its key role in the processing of the original spacer. On the other hand, Cas4 ensures that the CRISPR spacers are integrated in a specific direction, ultimately triggering the CRISPR immune response [15]. Taken together, these findings provide a key role for Cas4 nucleases in CRISPR arrays, providing for in situ spacer generation and functional compartment integration.

Figure 4. Mechanism of action of Cas4 [14-15]
2. Refrigeration electron microscopy technology to further analyze the working mechanism of CRISPR/Cas system <br> Although the CRISPR system has been widely used in various fields of biomedicine to biotechnology or synthetic biology, the delicate working mechanism is still lacking. Learn more. This year's three groundbreaking studies revealed the structure and working mechanism of different CRISPR systems through cryo-freezing electron microscopy, which provided an important theoretical basis for the improvement and clinical treatment of the CRISPR/Cas system. 1) Interpretation of the molecular mechanism of the type I CRISPR/Cas system : On July 6, 2018, researchers from Cornell University and Harvard Medical School published a paper in the journal Science to study the use of single-particle cryo-electron microscopy (cryo-EM). Analysis of the structure of the TfuCascade/R-loop/Cas3 ternary complex before and after R-loop cleavage reveals the confusing Cas3 recruitment, DNA cleavage and degradation mechanisms. These studies provide a structural basis for understanding the molecular mechanism of action of the Type I CRISPR/Cas system [16].
2) Cas13d functional structure study : On September 20, 2018, scientists at the Salk Institute in the United States used cryo-electron microscopy to analyze the structure and a series of Cas13d-sgRNA binary complexes and Cas13d-sgRN-target RNA ternary complexes. The dynamic process, which allows us to see in more detail how the Cas13d-system is guided to RNA and the process of cutting. This has improved the CRISPR system for us to lay the foundation for the treatment of RNA diseases more effectively [17].
3) Cas12d loop conformation analytical: December 13, 2018, researchers from Novo Nordisk Foundation Center using a cryogenic electron microscopy, clarified Cas12a targeting DNA cleavage and degradation of ssDNA random mechanism, revealing Cas12a cleaves its target DNA, releasing non-specific cleavage activity, and degrading ssDNA molecules after activation. This allows us to adjust the CRISPR engine to achieve a specific desired effect [18].

Figure 5. Cryogenic cryo-electron microscopy reveals the structure and working mechanism of the CRISPR system [16-18]
Fourth, breakthrough research on genetic editing based on CRISPR system The CRISPR/Cas gene editing system has great potential in human therapeutic applications. A full understanding of the repair mechanism of the in vivo editing system will help us to perform gene editing more efficiently and accurately, and provide sufficient theory and technology for safe and effective clinical applications. basis.
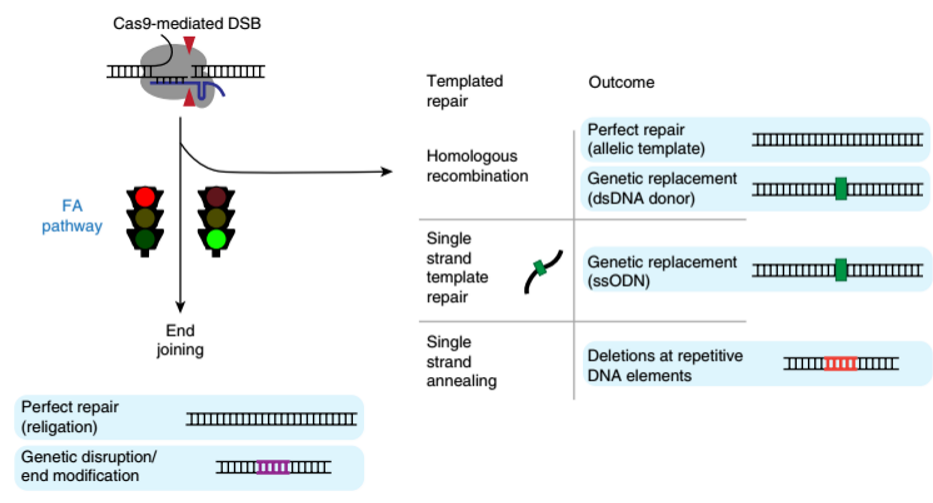
1. Fanconi anemia pathway plays a key role in the process of editing and forming DSBs in CRISPR-Cas9 editing
In August 2018, researchers at the University of California, Berkeley published a journal in the journal Nature Genetics, overturning the previous hypothesis that "cells repair genes after Cas9 enzyme cleavage of DNA". Researchers use CRISPR interference technology to 2000. Multiple genes were silenced and the FANCD2 protein in the Fanconi anemia pathway was localized to Cas9-induced DSBs, suggesting that it plays a key role in regulatory genome editing - regulation of FANCD2 protein can increase HDR frequency . At the same time, it was found that the Fanconi anemia pathway was not related to NHEJ, but the mechanism of repair to the single-chain template repair mechanism was improved by increasing the efficiency of homologous recombination. Therefore, the editing efficiency of HDR can be improved by regulating the activity of the Fanconi anemia pathway. This finding will help increase the efficiency of insertion of foreign DNA into the genome and ensure that CRISPR editing achieves the desired results [19].

Figure 6. Fanconi anemia (FA) pathway-mediated double-strand break (DSB) repair [19]
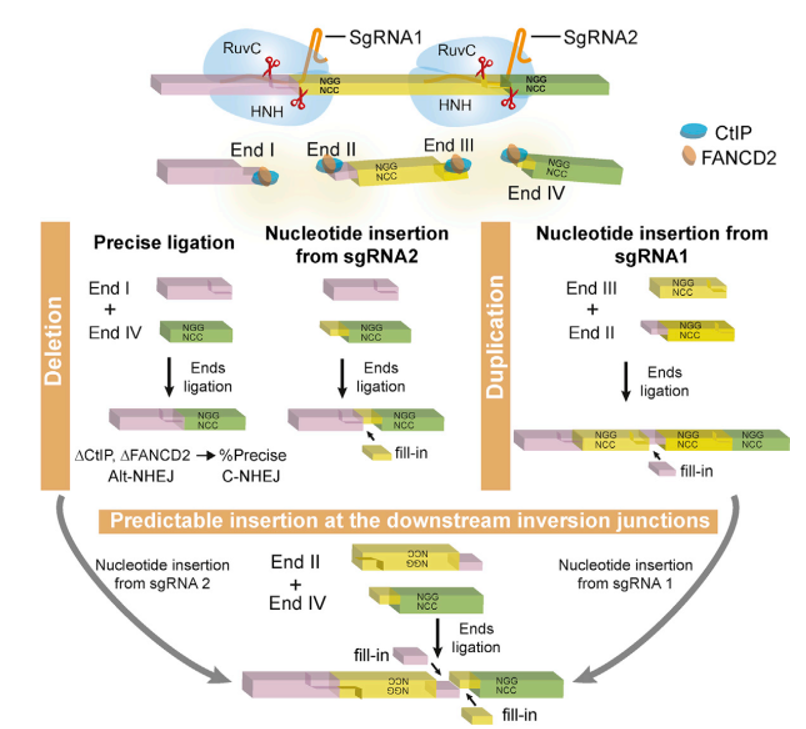
2. Accurate and predictable CRISPR chromosomal rearrangement reveals Cas9- mediated base insertion On July 19, 2018, Wu Qiang's research team from Shanghai Jiaotong University published the latest research results of the team in the Molecular Cell journal, subverting the existing gene editing theory. They found that the cis-cutting of Cas9-cleaved DNA produced a prominent end, not just the previously reported flat-end end form, but also used high-throughput sequencing to analyze the DNA repair methods of paired-directed RNA editing and found that the repair of the broken ends It is predictable and presents a very regular form. This disruptive discovery laid a solid foundation for optimizing and transforming gene editing techniques [20].

Figure 7. The repair of the CRISPR system after cutting is accurate and predictable [20]
3. The accuracy of templateless CRISPR/Cas9 gene editing is revealed. <br> For a long time, after stencil is not provided, the repair after CRISPR cleavage is generally considered to be random and heterogeneous. On November 7, 2018, in the Nature Journal, the HMax W. Shen team found that more than 2000 sites in the genome of CRISPR/Cas9 were used to detect how cells were repaired. The results showed that the template-free Cas9 gene editing was predictable. The edited gene does not contain a large number of variations, but a single result. At the same time, by introducing these big data, they established a machine learning model (indelphi), which can predict the repair results of gene editing in five human and mouse cell lines with high precision, and use this prediction to test human diseases. Repair effect. This model accurately corrects disease-causing genotypes to predicted genotypes, enabling accurate correction of disease-related mutations in humans. This study established the basis for accurate, template-free genome editing [21]. Subsequently, on November 27, 2018, the Felicity Allen team also published a report in Nature. They synthesized more than 40,000 pairs of guided RNA and targeted DNA sequence editing library systems to explore the cutting and repair mechanism of CRISPR/Cas9, and clarified the results of the repair. Depending on the DNA sequence of the targeting region, most reproducible mutations are the result of single base insertions, short deletions or longer micro-homology-mediated deletions. At the same time, a machine learning algorithm (FORECasT) was developed using Big Data, which can be used to predict the corrected results using only the target site DNA sequence [22]. The in-depth understanding of these mechanisms and the development of new tools will help people better design genetic editing experiments.
In summary, we briefly introduced the important events in the development and mechanism research of the new CRISPR system in 2018. At present, CRISPR-Cas9 has become a simple, accurate and rapid gene editing technology, which is widely used in many life science related fields such as medicine, agriculture, forestry and animal husbandry, such as eye diseases, blood diseases and other genetic diseases. At the same time, we also recognize that this technology still has many problems and many potentials to be further researched and expanded. We believe that in the near future, CRISPR-Cas9 will show its invaluable value in various fields.
references:
1. 201302_Science_Multiplex Genome Engineering Using CRISPR-Cas Systems
2. Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13[J]. Science, 2017: eaaq0180.
3. Konermann, Silvana, et al. “Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors.†Cell (2018) pii: S0092-8674(18)30207-1.
4. Athukoralage JS, Rouillon C, Graham S, et al. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate [J]. Nature, 2018.
5. Lucas B. Harrington1,*,†, David Burstein2,*,‡, Janice S. Chen et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science, 2018.
6. Zetsche B, Gootenberg J, Abudayyeh O, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System [J]. Cell, 2015: S0092867415012003.
7. Winston X. Yan et al. Functionally diverse type V CRISPR-Cas systems, 2018, doi:10.1126/science.aav7271.
8. Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities[J]. Nature, 2015, 523(7561):481-485.
9. Doron S, Melamed S, Ofir G, et al. Systematic discovery of antiphage defense systems in the microbial pangenome [J]. Science, 2018: eaar4120.
10. Hu JH , Miller SM , Geurts MH , et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity [J]. Nature.
11. Hiroshi Nishimasu, Xi Shi, Soh Ishiguro et al. Engineered CRISPR/Cas9 nuclease with expanded targeting space. Science, 21 Sep 2018, 361(6408):1259-1262, doi:10.1126/science.aas9129.
12. Pranam Chatterjee1,2,*,*,terjee1,2,*,1,2,*,ered CRISPR/Cas9 nuclease with expanded targeting space. Science, 21 Sep 20ortholog. Science Advances, 24 Oct 2018, 4(10) :eaau0766.
13. Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation[J]. Science, 2014, 343(6176):1247997.
14. Sebastian N. Kieper, Cristóbal Almendros, Juliane Behler et al. Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell Reports, 27 March 2018, 22(13):3377–3384, doi:10.1016/j.celrep. 2018.02.103.
15. Masami S, Garrett SC, Graveley BR, et al. Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci [J]. Molecular Cell, 2018, 70(5): 814-824.e6 .
16. Yibei X, Min L, Dolan AE, et al. Structure basis for RNA-guided DNA degradation by Cascade and Cas3 [J]. Science, 2018: eaat0839.
17. Zhang C, Konermann S, Brideau NJ, et al. Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d [J]. Cell, 2018, 175(1): 212-223.e17.
18. Stefano Stella, Pablo Mesa, Johannes Thomsen, et al. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity, Cell (2018). DOI: 10.1016/j.cell.2018.10.045.
19. Chris D. Richardson, Katelynn R. Kazane, Sharon J. Feng et al. CRISPR–Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nature Genetics, August 2018, 50(8):1132–1139, doi :10.1038/s41588-018-0174-0.
20. Precise and Predictable CRISPR Chromosomal Rearrangements Reveal Principles of Cas9-mediated Nucleotide Insertion
21. Max W. Shen et al., Predictable and precise template-free CRISPR editing of pathogenic variants, Nature, 2018, DOI: 10.1038/s41586-018-0686-x.
22. Felicity Allen et al, Predicting the mutations generated by repair of Cas9-induced double-strand breaks, Nature Biotechnology (2018). DOI: 10.1038/nbt.4317.
Diagnosis Device,Medical Diagnosis Devices,Diagnosis Therapy Device,Health Diagnosis Device
Medton Medical , https://www.medton.cn